- Overview
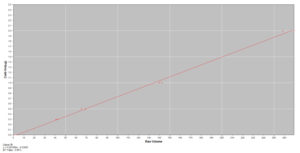
- STD Curve
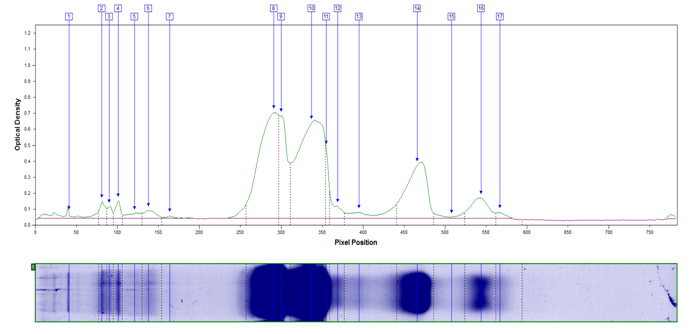
- Relative Dye Binding
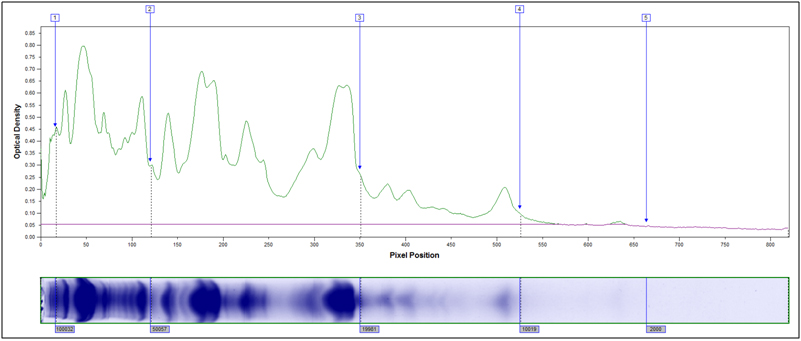
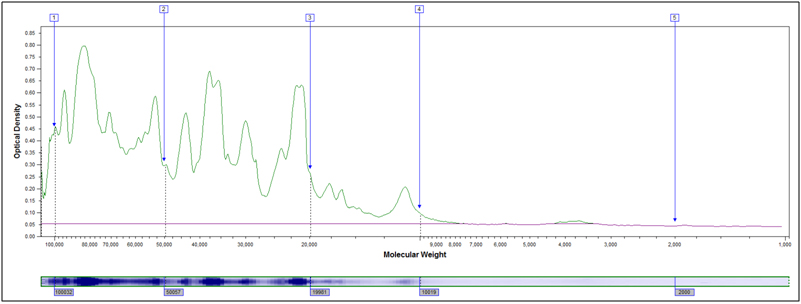
- MW Distribution
Overview
Food & Nutritional Protein Analysis
Clients from the food, dairy, supplement, personal care, and other industries rely on Kendrick Labs’ 1D gel electrophoresis testing service to analyze proteins in their products.
We offer two methods for quantification of proteins resolved by 1D: Relative Dye Binding or Percentage by Weight. Either method can be used to compare differences between samples; results are expressed in different units.
Relative Dye Binding Protein Quantification: Determine relative amount of proteins present in a sample. Example Relative Dye Binding Report (PDF)
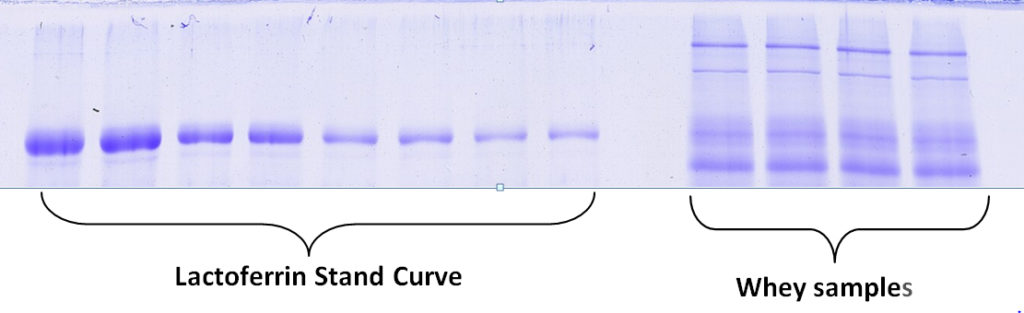
Percentage by Weight Protein Quantification: Determine percentage by weight of an individual protein present in a sample compared to a standard. Example Percentage by Weight Report (PDF)
We can also do:
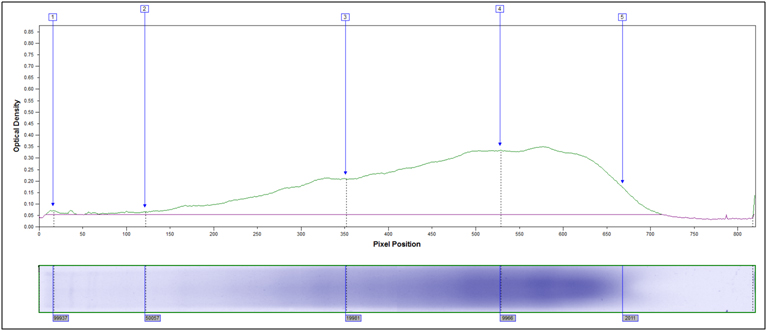
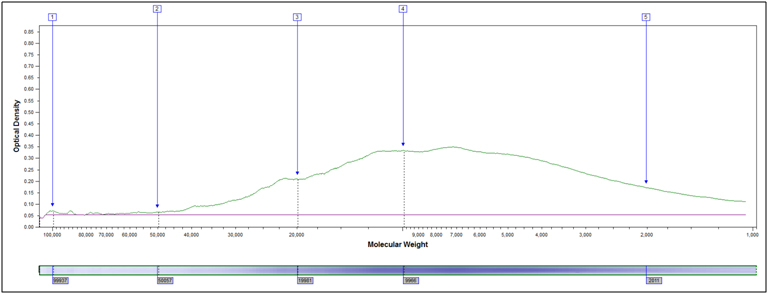
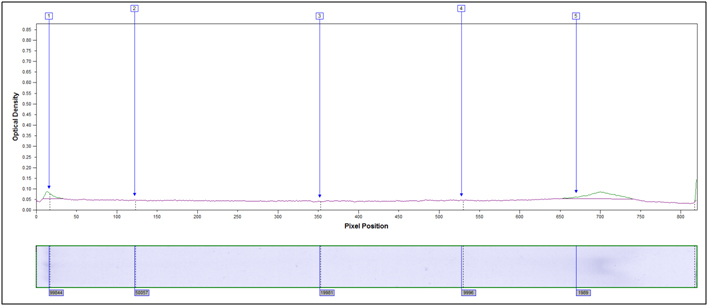
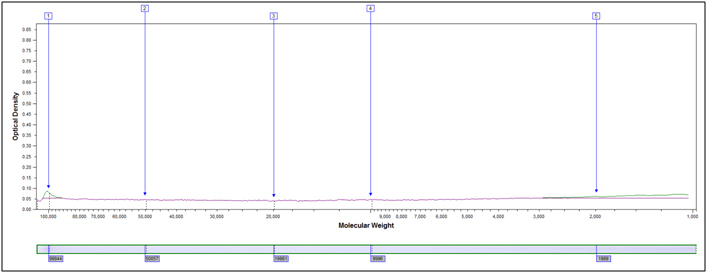
Molecular Weight Distribution: Determine the size (molecular weight) of proteins present in your sample. Example MW Distribution Report (PDF)
Custom Analysis: Contact us to discuss your project, and our experts will create a customized plan for you.
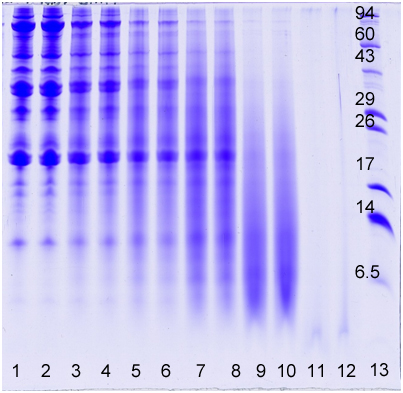
How does 1D gel electrophoresis work? 1D Electrophoresis (aka SDS PAGE) is useful for separating proteins in a mixture by size. Below is an image of a Coomassie blue-stained gel loaded with common food samples to show the different protein banding patterns
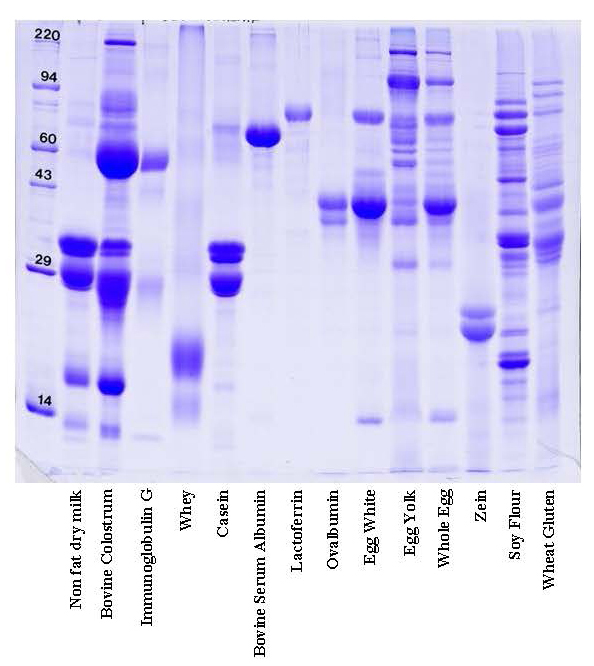
In this method, samples are measured, solubilized and denatured in an SDS-containing buffer, and then separated by size in a gel matrix. Proteins in the gel are stained with Coomassie blue to visualize them. Molecular weight standards allow for calibration of protein size. Proteins can be quantified by determining band percentage (stain density of each protein as a % of total stain density of all proteins in the lane) or percentage by weight of the sample (quantified relative to a standard curve of the protein of interest). In addition, individual bands can be cut out of the gel, and proteins identified by Mass Spectrometry.
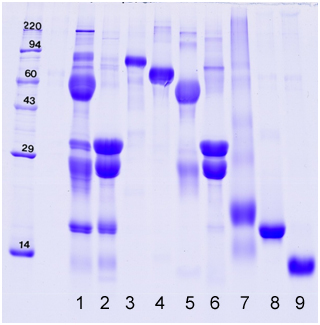
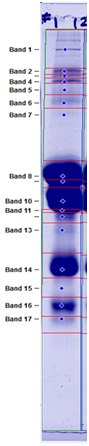 Band/Protein/Percentage
Band/Protein/Percentage